Christopher Rae
I’m interested in the mycobacterial cell wall – a unique and important interface between host and pathogen. Using genetics, biochemistry, and structural biology I am studying how cell wall permeability is critical for virulence of Mycobacteria.
Depicted on the right: Structure of a periplasmic channel from Mycobacterium smegmatis.

Zoe Netter
I am exploring the interplay between mycobacterial pathogenesis and defense against phages (viruses of bacteria) and other mobile genetic elements. Human immune responses to mycobacterial infections are becoming more well-defined, but very little is known about the immune systems of the pathogens themselves and how they have evolved to survive under the threat of phage predation. The lifestyles of the diverse and ubiquitous phages that infect mycobacteria are also relatively unknown. I am developing genetic tools to assess determinants of mycobacterial phage infection and to identify trade-offs and strategies that mycobacterial pathogens employ to balance virulence activity with phage defense.
Rita McCall
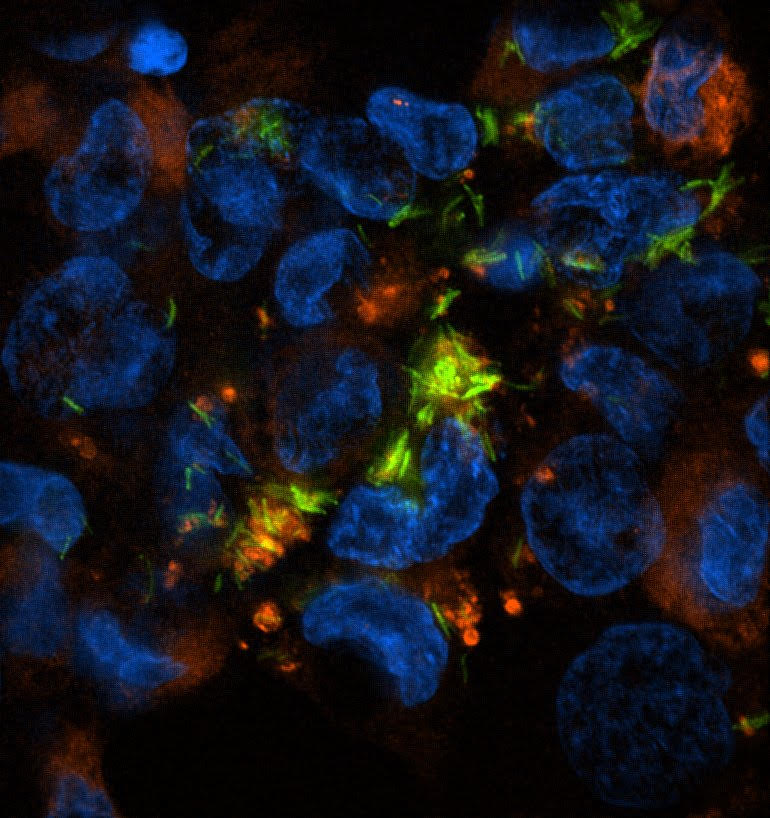
I love thinking about what is happening inside a macrophage during Mycobacteria tuberculosis (Mtb) infection. I’m fascinated by the complex host response induced upon Mtb’s perforation of the phagosome. I use various “omics” and more targeted genetic approaches to uncover novel aspects of the macrophage’s response to infection.
Depicted on the left: human derived THP-1 cell line infected with M. tuberculosis (in green) colocalized with host protein Galectin-8 (in red). Blue is a DAPI stain showing the nucleus of individual cells.
Nick Garelis
I am interested in investigating how host immune responses can increase susceptibility to Mycobacterium tuberculosis (Mtb), using CRISPR screens to identify host factors that may be important for control of Mtb, and in understanding the mechanisms underlying mycobacterial secretion systems and the trafficking of mycobacterial outer membrane proteins.
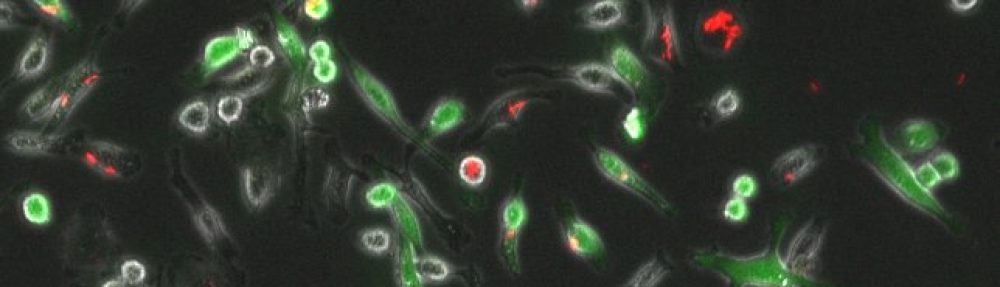
Depicted on the right: different strains I’ve made for studying mycobacterial pathogenesis.

Ellie Reinhart
I am exploring how macrophages restrict the growth and virulence of Mycobacterium tuberculosis (Mtb). We know that if you remove the ESX-1 secretion system of Mtb, mutants fail to kill macrophages or mice and also fail to grow at early time points. However, it is unknown how the host is able to restrict mutant bacterial growth and cytotoxicity. To elucidate the mechanism of action, I use CRISPR screens, classic genetic approaches and immunofluorescent microscopy techniques.
Christine Qabar
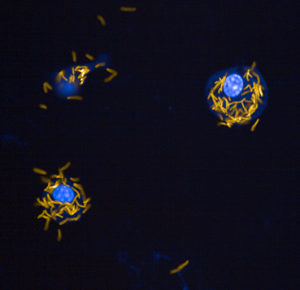
I am fascinated by the ability of Mycobacteria to survive and thrive in the face of environmental challenges, like low oxygen, nutrient starvation, and reactive oxygen/nitrogen species. Using bacterial genetics techniques, I seek to understand what makes these intracellular pathogens so resistant in the face of powerful immune strategies.
Depicted on the left: Mouse bone marrow-derived macrophages (blue) infected with a strain of Mycobacterium marinum (yellow) lacking one metabolic pathway.
Sydnee T. Gould
I work on identifying bacterial genes responsible for Mycobacteria tuberculosis (Mtb)’s ability to enter and survive in the brain. Though commonly a respiratory disease, some Mtb strains have demonstrated heightened ability to infiltrate the brain in children. I am interested in what host or bacterial factors make particular strains better suited to cross the blood-brain-barrier and proliferate. To learn more about this phenomenon, I use wild mice that have been shown to have heightened susceptibility to Mtb meningitis.

Laura Britt
I focus on Mycobacterium avium (M. avium) and two distinct colony morphologies which heavily correlate with virulence and antibiotic resistance. I am researching the mechanism of reversible switching between the two morphotypes to understand how and why these two cell programs exist using a variety of genetic techniques.
Depicted on the left: Smooth opaque (SmO) and smooth transparent (SmT) colonies of M. avium.